Here is a term paper on ‘Recombinant DNA Technology’. Find paragraphs, long and short ‘Recombinant DNA Technology’ especially written for school and college students.
Term Paper on Recombinant DNA Technology
Term Paper Contents:
- Term Paper on the Meaning of Recombinant DNA Technology
- Term Paper on the Concept of Recombinant DNA Technology
- Term Paper on the Goals of Recombinant DNA Technology
- Term Paper on the Types of Recombination DNA Technology
- Term Paper on the Technique of Recombinant DNA Technology
- Term Paper on the Tools Required for Recombinant DNA Technology
- Term Paper on the Application of Recombinant DNA Technology
Term Paper # 1. Meaning of Recombinant DNA Technology:
The technique of removing, modifying or adding genes to a DNA molecule (of an organism) in order to change the information it contains is called Recombinant DNA (rDNA) technology.
By changing this information, genetic engineering changes the type or amount of proteins an organism is capable of producing, which enables us to make new substances or perform new functions. The main and constructive objective of genetic engineering is to produce desired characteristics and to eliminate undesirable ones.
Recombinant DNA (rDNA) technology is a field of molecular biology in which scientists ‘edit’ DNA to form new synthetic molecules. The practice of cutting, pasting and copying DNA dates back to Arthur Kornberg who made successful replication of viral DNA. This was followed by the Swiss biochemist Werner Arber’s discovery of restriction enzymes in bacteria that degrade foreign viral DNA molecules while sparing their own DNA.
Arber effectively showed geneticists how to ‘cut’ DNA molecules; which was followed by the understanding that ligase could be used to ‘glue’ them together. These two achievements launched rDNA technology research and allowed man to ‘play god’ for the first time in human history. In 1978, scientists were able to replicate somatostatin, the protein that regulates human growth hormones.
Term Paper # 2. Concept of Recombinant DNA Technology:
Recombinant DNA technology includes techniques of cutting DNA into specific fragments using enzymes restriction endonucleases and joining the fragments with the help of enzyme, ligase. Using this technology, the fragments of foreign DNA can be inserted into a vector which may be plasmids or viruses. This technology bypasses the restriction in the gene transfer mechanisms between unrelated organisms.
Recombinant DNA technology or genetic engineering involves the following steps:
(i) DNA fragments coding for proteins of interest are synthesized chemically or isolated from an organism.
(ii) These DNA fragments are inserted in a restriction endonuclease cleavage site of the vector that does not inactivate any gene required for vector’s maintenance.
(iii) The recombinant DNA molecules are now introduced into a host to replicate.
(iv) Recipient host cells that have acquired the recombinant DNA are selected. Selection pressure is applied to enrich bacteria with a selectable marker.
(v) Desired clones are then characterised to ensure that they maintain true copies of the DNA segment that was originally cloned.
Term Paper # 3. Goals of Recombinant DNA Technology:
The goal of recombinant DNA (rDNA) technology is to introduce recombinant genes into a host cell along with expression factor, so that the host cell expresses the desired protein. For this process, scientists need to ‘turn off’ the signals that tell a host cell to destroy or degrade the genes being introduced.
Recombinant DNA research is a challenging field and holds great promise for the future. In the future, rDNA technologies will play a key role in producing targeted medicines, preventing genetic diseases, and providing patients with less toxic pharmaceuticals. It will also impact agriculture and livestock as the researchers find ways to optimise the genetic codes of plants and animals to resist diseases.
So far the global sale of recombinant pharmaceutical products has been very impressive with a total value of Rs. 21253 billion in the year 1996. In India, nine products have been approved for marketing, a few of which are as follows – alpha interferon (cancer drug), insulin (diabetes drug), hepatitis B vaccine, blood clotting factor 7, erythropoietin (drug used in kidney failure), streptokinase (drug given in heart attack), and human growth hormone. All these products except hepatitis B vaccine are being imported. In 1996-97, the total import of these drugs was about 237 crores.
Term Paper # 4. Types of Recombination DNA Technology:
The following three types are most commonly observed:
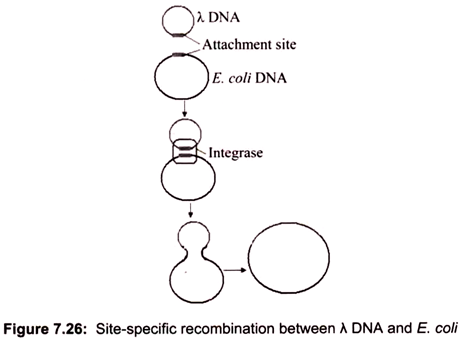
i. Site-Specific Recombination:
Site-specific recombination occurs at a specific DNA sequence. The first example was found in the integration between λ DNA and E. coli DNA. Both of them contain a sequence, 5′-TTTATAC-3′, called the attachment site, which allows the two DNA molecules to attach together by base pairing. Once attached, the enzyme integrase catalyzes two single strand breaks as in the Holliday model. After a short branch migration, the integrase exerts a second strand cuts on two other strands (Fig. 7.26). Resolution of two Holliday junctions completes the integration process.
ii. Homologous Recombination:
Homologous recombination occurs between two homologous DNA molecules. It is also called DNA crossover. During meiosis, two homologous pairs of sister chromatids align side by side (Fig. 7.27). The DNA crossover is very likely to occur. It could be as often as several times per meiosis. The mechanism of DNA crossover was first explained by Robin Holliday in 1964.
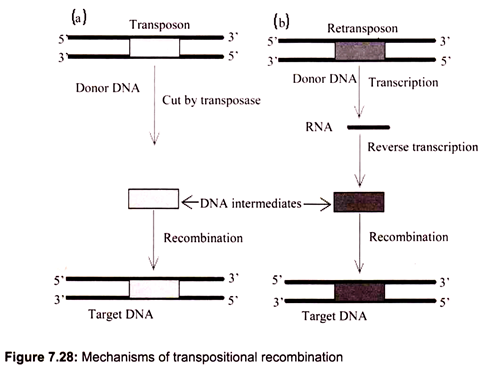
iii. Transpositional Recombination:
Transpositional recombination is a process in which a mobile element is inserted into a target DNA. It may occur by one of two mechanisms such as directly as DNA or through RNA (Fig. 7.28). The mobile elements that transpose through DNA are called transposons and those via RNA are referred to as retrotransposons.
In bacteria, the target DNA is cut by transposase, producing sticky ends (single strands, easy to pair with complementary sequences). The transposase also has ligase activity which may ligate the intermediate DNA of transposons. The gaps at sticky ends are filled by DNA polymerase, generating direct repeats. Other organisms are expected to use the same mechanism to insert the DNA intermediates of either transposons or retrotransposons into target DNA. The direct repeats generated by this mechanism are found in all mobile elements.
(a) For Trasposons:
The transposon in donor DNA is cut by a special enzyme and then inserted into a target DNA. In bacteria, this enzyme is called transposase which has both nuclease and ligase activities.
(b) For Retrotransposons:
The retrotransposon in donor DNA is first transcribed into RNA and then reverse-transcribed into DNA, which is inserted into a target DNA by the same recombination mechanism as the DNA intermediates of transposons.
Term Paper # 5. Technique of Recombinant DNA Technology:
The technique of recombinant rDNA technology involves three stages.
Stage I – Isolation:
It involves the purification of DNA from the donor organism and isolation of fragments, one of which will contain the required gene. After isolation, the process of cloning makes multiple copies of this gene. Gene of interest is then identified.
Stage II – Insertion of Gene into Vector:
Isolated gene is introduced into the vector, either a bacterial plasmid or virus, after this multiple copies of gene are produced in the vector.
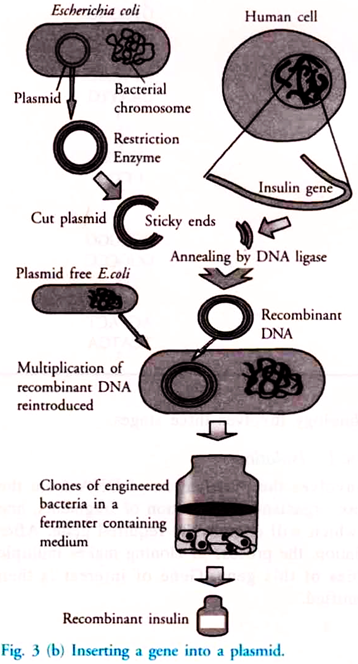
To insert a piece of DNA into a plasmid, the plasmid is cut open, using a restriction endonuclease. Restriction endonuclease makes staggered cuts in the DNA, leaving a short length of unpaired base at each end.
These are called sticky ends DNA ligase is used to join the sugar – phosphate backbone of the plasmid and the inserted DNA together. This new DNA molecule – part of plasmid DNA and part of original DNA is called recombinant DNA [Fig. 3(a)].
Stage – III:
Vector inserts gene into recipient organism, where it expresses itself.
After the above steps it is essential that the cell starts making the gene product. Now an economical method is to be devised for its mass production. Fig. 3(b) shows different steps involved in the production of human insulin.
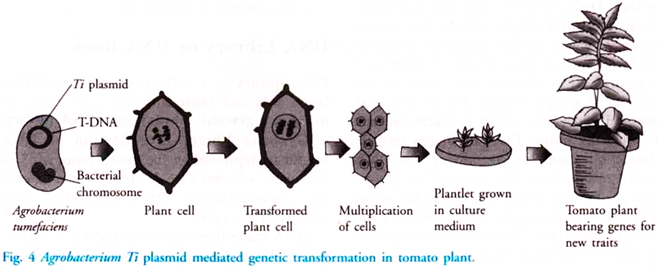
Other than production of human insulin tumor – inducing plasmid is another example of rDNA technology. This occurs in the soil bacterium Agrobacterium tumefaciens. Tomato, soyabean, cotton and sunflower are the broad- leaf crops that are infected by this bacteria. This bacterium is responsible for the formation of cancerous growth called crown gall tumour.
This phenomenon is seen because of the presence of Ti plasmid present in pathogenic bacterium. For the purpose of genetic engineering, Agrobacterium strains in which the tumour forming gene has been deleted, is developed. These bacteria can still infect plant cells. Part of Ti plasmid transferred into plant cell DNA, is called as T-DNA.
This T-DNA along with the desired DNA is spliced and inserted into the chromosomes of the host plant where it produces copies of its kind. Now it does not produce tumours. Plant cells with this T-DNA are cultured, induced to multiply and differentiated to form plantlets. These plantlets are then transferred to soil where they grow into plants (Fig. 4). These plants have a foreign gene, which expresses itself.
Term Paper # 6. Tools Required for Recombinant DNA Technology:
Recombinant DNA technology can be used to generate valuable polypeptides, insulin, interferons, growth hormones and control genetic diseases, etc. Certain biological tools are required to carry on the process of rDNA.
1. Enzymes:
There are certain enzymes, which help in rDNA technology.
These are as follows:
i. Exonuclease:
These enzymes act on genome and digest the base pair on 5’ or 3’ ends of the single DNA strand or at single strand nicks or gaps in double strand DNA.
ii. Endonuclease:
These enzymes act on genetic material and cleave the double stranded DNA at any point except the ends. This action only takes place on one strand of DNA.
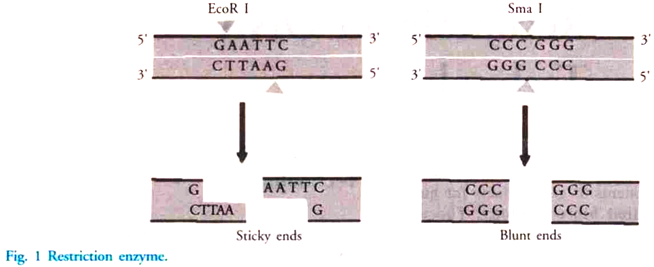
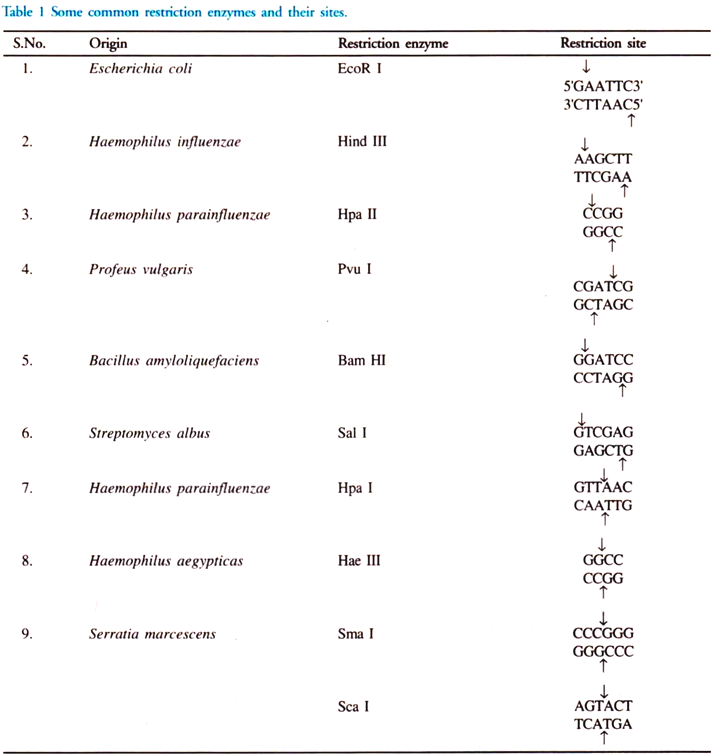
iii. Restriction Endonuclease:
These are enzymes that cleave the sugar-phosphate backbone of DNA. A given enzyme of this type cuts both strands of duplex DNA within a stretch of just a few bases. They are naturally found in bacteria as a chemical weapon against invading viruses. These enzymes act on both the strands of DNA when certain foreign nucleotides are introduced into the cell.
There are two types of restriction endonuclease, type I and II. Type I enzyme cuts DNA at random sites that can be more than 1000 base pairs away from the recognized sequence. Type II restriction endonuclease is simple and requires no ATP for degradation of DNA. They cut DNA within the recognition site (Fig. 1).
iv. DNA Ligase:
Mertz and Davis (1972) were the first to demonstrate that cohesive termini of cleaved DNA molecules could be covalently sealed with E.coli DNA ligase and produce recombinant DNA molecules. DNA ligase helps in sealing single strand nicks in DNA which have 5’→ 3’ -OH (hydroxyl) termini. The two DNA ligase extensively used are the ligase from E.coli and that encoded by T4 phage, hence the enzyme is also known as T4 DNA ligase.
v. Reverse Trancriptase:
This enzyme is used to synthesise a complementary DNA (cDNA) by using mRNA as a template.
vi. DNA Polymerase:
This enzyme polymerises the DNA synthesis on DNA template.
2. Foreign DNA/Passenger DNA:
The gene of interest is identified on a gemone and pulled out. This is known as foreign or passenger DNA which is a fragment of DNA molecule. It is enzymatically isolated and cloned. The cloned DNA expresses normally as in parental cell.
3. Cloning Vector:
Vectors are those DNA molecules that can carry off foreign DNA fragments when inserted into it. Vector can also be called as vehicle DNAs. Vectors are grouped into the following categories – bacterial plasmids, bacteriophages, Yeast artificial chromosomes, Bacterial artificial chromosome and plasmids.
i. Plasmids:
These are extra chromosomal, double stranded, closed, circular and self-replicating DNA molecules present in the bacterial cell. Plasmids contain enough genetic information for their own replication (Fig. 2).
ii. Cosmid and Phagemid:
Cosmid is a recombinant plasmid having a DNA sequence of bacteriophage lamda required for packaging. It allows cloning of large DNA fragments. Phagemid is formed by recombining a phage with plasmid.
iii. Bacteriophages:
A bacteriophage is a virus which attacks a bacteria. These are required for cloning a large DNA fragment.
iv. Yeast Artificial Chromosomes (YAC):
This vector is capable of carrying a large fragment of DNA (up to 2 Mb). The only disadvantage is that its transformation efficiency is very low. In this target, DNA is partially digested with the help of restriction enzyme and the same enzyme also cleaves YAC. The cleaved YAC is ligated with a digested DNA fragment to form an artificial chromosome (Fig. 2).
v. Bacterial Artificial Chromosome (BAC):
A vector used to clone DNA fragments in E.coli cells.
Term Paper # 7. Applications of Recombinant DNA Technology:
Different applications of recombinant DNA technology is shown in Table 1.
i. Human Insulin:
Treatment of insulin-dependent diabetes.
ii. Human Growth Hormones:
Replacement of missing hormones in short stature people.
iii. Clacitonin:
Treatment of rickets
iv. Chronic Gonadotropin:
Treatment of infertility.
v. Blood Clotting Factor VIII/IX:
Replacement of clotting factors missing in patients with hemophilia A/B.
vi. Tissue Plasminogen Activator:
Dissolving blood clots after heart attacks and strokes.
vii. Erythropiotin:
Stimulation of the formation of erythrocytes (RBCs) for patients suffering from anaemia during kidney dialysis or side effects of AIDS patients treated by drugs.
viii. Platelet:
Stimulation of wound healing
ix. Interferon:
Treatment of pathogenic viral infections, cancer.
x. Interleukins:
Enhancement of action of immune system.
ci. Vaccines:
Preventions of infectious diseases such as hepatitis B, herpes, influenza, pertussis, meningitis etc..