Applications of Microarrays in Various Fields!
Microarrays used to monitor the expression level of genes in comparison between two conditions remain one of the most widespread uses of microarrays. This type of study, termed gene expression profiling, can be used to determine the function of particular genes during a particular state, such as nutrition, temperature, or chemical environment.
Such results could be observed as up- or down-regulation, or unchanged during particular conditions.
For example, a group of genes could be up- regulated during heat shock, and as a group, these genes could be assigned as heat shock responsive genes. Some genes in this group may have already been identified as heat shock responsive, but other genes in the group may not have been assigned any function.
Based on a similar response to heat shock, new functions are then assigned to the genes. Therefore, extrapolation of function based on common changes in expression remains one of the most widespread applications of micro-arrays.
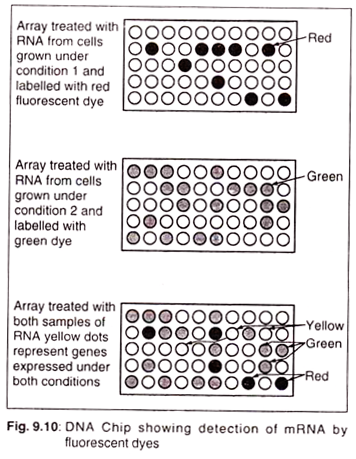 It is also used to find out which genes are expressed under which conditions. DNA chips can monitor many different mRNAs at one time. Each spot on the grid has a different DNA sequence attached. To determine which genes are expressed under which conditions, mRNA is isolated and each sample is labelled with a different fluorescent dye.
It is also used to find out which genes are expressed under which conditions. DNA chips can monitor many different mRNAs at one time. Each spot on the grid has a different DNA sequence attached. To determine which genes are expressed under which conditions, mRNA is isolated and each sample is labelled with a different fluorescent dye.
If two different dyes are used, the same chip can be used for both.
It is then visualized in three different ways: one shows only the red dye, another only the green, and the third merges the two images so overlapping spots look yellow:
1. In agriculture, microarrays have been used to identify genes which are involved in the ripening of tomato’s, for example. In this type of study, RNAs are isolated from raw and ripened fruit, and then compared to determine which genes are expressed during the process. Genes which are down-regulated during the ripening may also provide useful information about the process.
2. On a basic scientific level, microarrays have been used to map the cellular, regional, or tissue-specific localization of genes and their respectively encoded proteins.
3. Microarrays have been used at the subcellular level to map genes that encode membrane or cytosolic proteins; at the cellular level to map genes that distinguish between different types of immune cells; at the tissue region level to distinguish genes which encode hippocampus or cortex brain region specific proteins; and at the tissue level to identify genes which are expressed in muscle, liver, or heart tissues.
4. Pharmacological studies have also used microarrays as a means of discerning the mechanism of action of therapeutic agents and as a corollary to develop new drug targets. The guiding principle in this endeavour is that genes regulated by therapeutic agents result from the actions of the drug.
Identification of the genes that are regulated by a certain drug could potentially provide insight into the mechanism of action of the drug, prediction of toxicologic properties, and new drug targets.
One of the most exciting areas of application is the diagnosis of clinically relevant diseases. The oncology field has been especially active and to an extent successful in using microarrays to differentiate between cancer cell types.
The ability to identify cancer cells based on gene expression represents a novel methodology that has real benefits.
In difficult cases where a morphological or an antigen marker is not available or reliable enough to distinguish cancer cell types, gene expression profiling using microarrays can be extremely valuable:
A very recent application of microarrays has been to perform comparative genomic analysis. Genome projects are producing sequences on a massive level, yet there still do not exist sufficient resources to sequence every organism that seems interesting or worthy of the effort.
Therefore, microarrays have been used as a short cut to both characterize the genes within an organism (structural genomics) as well as to determine whether those genes are expressed in a similar way to a reference organism (functional genomics).
A good example of this is in the species Oryza sativa (rice). Microarrays based on rice sequences can be used to hybridize cDNAs derived from other plant species such as corn or barley. The genome sizes in the latter are simply too large for whole genome projects, so hybridization with microarrays to rice genes presents an agile way to address this question.
Indeed, the use of microarrays to determine whether a gene is present and whether it goes up or down under certain conditions will continue to spawn even more applications that now depend only upon the imagination of the microarray researcher.
