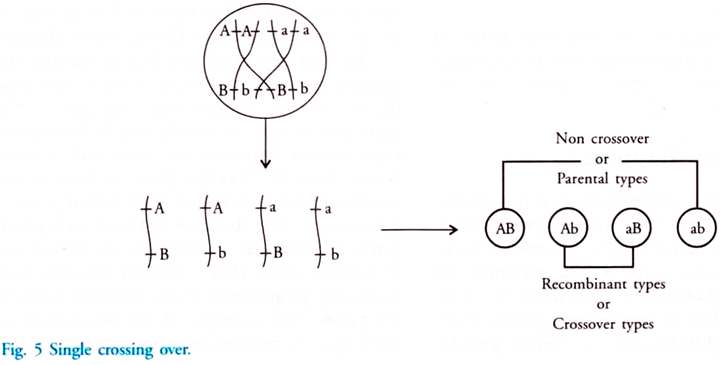
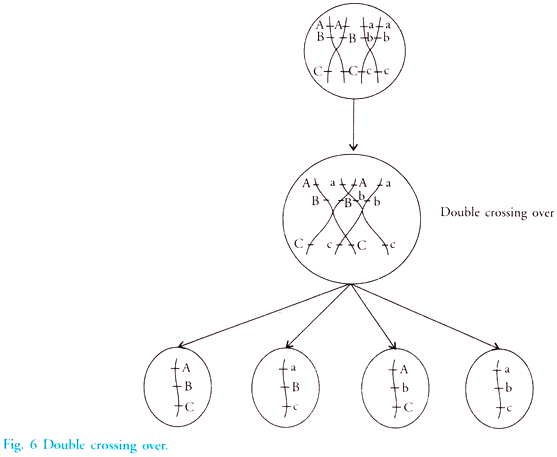
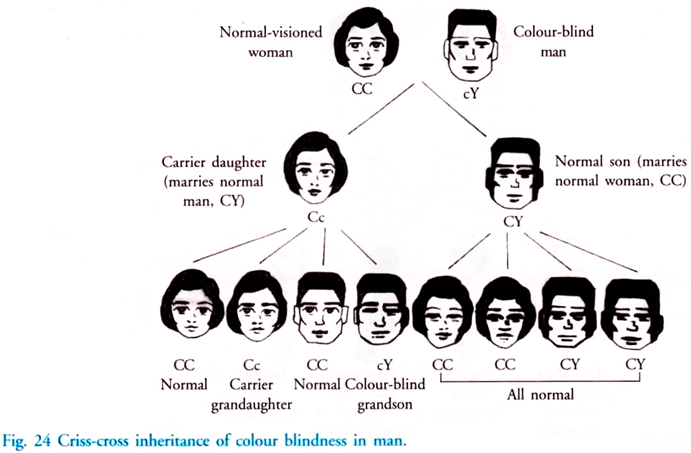
In this article we will discuss about DNA isolation with its protocol. Learn how to isolate:- 1. Plasmid DNA 2. Bacterial Genomic DNA 3. Yeast Genomic DNA 4. Genomic DNA from Blood 5. DNA from Animal Cells 6. Genomic DNA from Eukaryotic Tissues 7. Plant DNA using CTAB Extraction Method 8. Chloroplast DNA 9. Mitochondrial DNA.
Contents:
- How to Isolate Plasmid DNA?
- How to Isolate Bacterial Genomic DNA?
- How to Isolate Yeast Genomic DNA?
- How to Isolate Genomic DNA from Blood?
- How to Isolate DNA from Animal Cells?
- How to Isolate Genomic DNA from Eukaryotic Tissues?
- How to Isolate Plant DNA using CTAB Extraction Method?
- How to Isolate Chloroplast DNA?
- How to Isolate Mitochondrial DNA?
1. How to Isolate Plasmid DNA
?
Cohen and Boyer (1973) carried out the first cloning experiment. They took advantage of two developments. One was the isolation of enzymes known as restriction enzymes which could cut DNA at specific and predictable locations.
The second was the ability to link individual molecules resulting from restriction digestion using the enzyme ligase. Thus, a genomic DNA segment could be inserted into a self-replicating extra-chromosomal DNA from a bacterium and it could be used to grow many copies or clones of the cloned genomic DNA segment.
Isolation of plasmid DNA from E. coli is the most common and routine procedure in research laboratories. Plasmids are double-stranded circular DNA molecules that have the property of self-replication, independent of bacterial chromosomal DNA.
Though the presence of a plasmid in a bacterial cell is detected genetically as a change in phenotype of the bacteria, it is often necessary to isolate plasmid DNA for molecular studies such as size determination, restriction enzyme mapping, nucleotide sequencing or for construction of new hybrid plasmids, PCR, expression of proteins, transfection and gene therapy.
The plasmid DNA of the bacteria are closed circular molecules of double stranded DNA that range in size from 1 kb to more than 200 kb. They are found in bacteria as additional hereditary units and replicated independently of bacterial chromosome.
However, the plasmids rely upon enzymes and proteins from their host for their successful transcription and replication. Plasmids are useful to bacterial cells because they can carry genes that code for enzymes which may be involved in resistance to antibiotics, toxins in non-ideal environmental conditions, or production of toxins within the bacteria itself.
There are many methods available to isolate plasmids from bacteria. The widely-practiced procedure is the alkaline lysis of cells. This protocol is often referred to as ‘mini-prep’, and yields fairly clean DNA, quickly and easily.
1. Inoculate the bacterial colonies (preferably a single colony from a bacterial plate) using a sterile loop into 5 ml of nutrient broth—Luria-Bertani broth or (LB) medium—with ampicillin or any other appropriate antibiotic and grow overnight, shaking vigorously (180 rpm) at 37°C.
2. Centrifuge the culture taken in microtubes using a microfuge for 1 minute at 10,000 rpm. Remove the supernatant and drain the tubes briefly on paper towel.
3. Repeat step 2 with the same tube, filling the tube again with more bacterial culture.
4. Add 0.2 ml of ice-cold Solution I to the bacterial pellet and re-suspend cells as much as possible using disposable microtip or inoculation loop.
5. Add 0.4 ml of Solution II, cap the tubes and invert several times gently. Keep the tubes at room temperature for 5 minutes.
6. Add 0.3 ml of ice-cold Solution III and invert the tubes several times gently. Incubate the tubes on ice for 10 minutes.
7. Centrifuge the tubes for 5 minutes at 10,000 rpm in a microfuge and transfer the supernatant into fresh tubes. Try to avoid taking any white precipitate during transfer. It is better to leave a little supernatant behind to avoid accidentally taking the precipitate.
8. Add 1.0 ml of isopropanol to the microfuge tubes and leave at room temperature for 2 minutes.
9. Centrifuge at 10,000 rpm for 2-5 minutes. Discard the supernatant liquid and drain the tubes on a paper towel.
10. Add 1 ml of ice-cold 70% ethanol. Mix by inverting several times. Centrifuge the tubes for 1 minute at 10,000 rpm. Discard the supernatant and drain the tubes on paper towel.
11. Allow the tubes to dry for 5 minutes. Add 50 μI of TE to the tubes and dissolve the plasmid DNA.
Note:
i. Antibiotic is required to provide a selective pressure for maintaining the plasmid within the bacterial cells. Without this selective pressure, the bacteria tend to lose the plasmid and not replicate the extra-chromosomal plasmid if it is not ‘needed’.
ii. The method includes steps (step 3 of the protocol) to increase the starting volume of cells so that more plasmid DNA can be isolated in a single preparation.
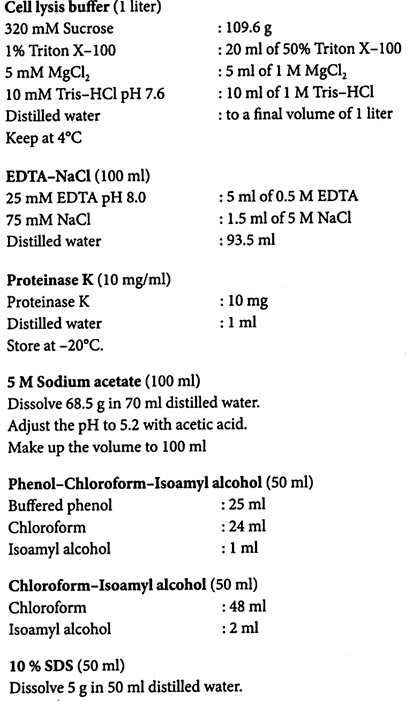
iii. Solution I contains glucose, Tris and EDTA. Glucose is added to increase the osmotic pressure outside the cells; Tris is a buffering agent used to maintain a constant pH (8.0); EDTA protects the DNA from degradative enzymes (DNases). EDTA binds divalent cations that are necessary for DNase activity.
iv. Solution II contains sodium hydroxide (NaOH) and sodium dodecyl sulfate (SDS; detergent). The alkaline mixture ruptures the cells and the detergent breaks apart the lipid membrane and solubilizes cellular proteins. NaOH also denatures the DNA into single strands.
v. Solution III contains a mixture of acetic acid and potassium acetate. The acetic acid neutralizes the pH, allowing the DNA strands to renature. The potassium acetate precipitates the SDS from solution along with cellular debris. The E. coli chromosomal DNA, a partially renatured tangle at this step, is trapped in the precipitate. The plasmid DNA remains in solution.
vi. The fractionation step (step 7 of the protocol) separates the plasmid DNA from the cellular debris and chromosomal DNA in the pellet.
vii. Isopropanol effectively precipitates nucleic acids, but is much less effective with proteins. A quick precipitation can therefore purify DNA from protein contaminants.
viii. The last fractionation step further purifies the plasmid DNA from contaminants.
ix. Ethanol helps to remove the remaining salts and SDS from the preparation.
Stock Solutions:
50% Glucose (200 ml):
Dissolve 100 g of glucose (dextrose) in 150 ml of distilled water. Make upto 200 ml and autoclave.
1M Tris-HCl pH 8.0 (500 ml):
Dissolve 60.55 g of Tris in 400 ml of distilled water. Adjust the pH to 7.5 with concentrated HCl. Make up the volume to 500 ml.
0.5 M EDTA pH 8.0 (100 ml):
Dissolve 16.8 g of anhydrous disodium salt of EDTA in 70 ml of distilled water. Adjust the pH to 8.0 using NaOH pellets. More EDTA will dissolve as the pH approaches 8.0. Make up the volume to 100 ml and autoclave.
10% SDS (100 ml) (w/v):
Dissolve 10 g in 100 ml distilled water. Heat gently to get SDS into solution.
1 N Sodium Hydroxide (100 ml):
Dissolve 4 g in 100 ml distilled water.
5M Potassium Acetate (500 ml):
Dissolve 245.5 g in 400 ml distilled water. Adjust the pH to 5.5 with glacial acetic acid. Make up the volume to 500 ml.
2. How to Isolate
Bacterial Genomic DNA?
The isolation of DNA from bacteria is a relatively simple process. The organism to be used should be grown in a favorable medium at an optimal temperature, and should be harvested in ‘late log’ to ‘early stationary’ phase for maximum yield.
The cells can then be lysed and the DNA isolated. Following lysis, the cellular constituents are selectively removed. Once this is accomplished, DNA can then be precipitated from solution with alcohol and dissolved in an appropriate buffer.
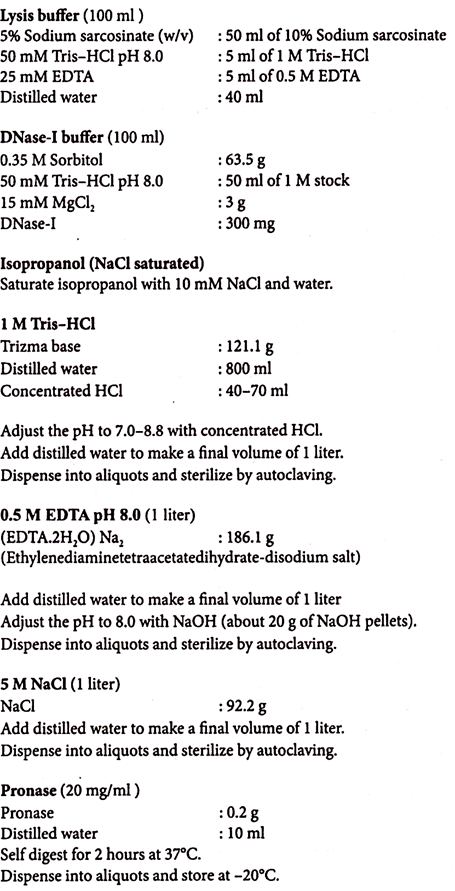
The lysis of the bacteria is initiated by re-suspending the bacterial pellet in a buffer containing lysozyme and EDTA. The EDTA disrupts the outer membrane of the gram-negative envelope by removing the Mg2+ from the lipopolysaccharide layer and additionally inhibits DNases.
This allows the lysozyme access to the peptidoglycan. After partial disruption of the peptidoglycan by the enzyme, a detergent such as SDS is added to lyse the cells. Most gram-negative cells are lysed after this treatment and many can even be lysed without lysozyme. Once the cells are lysed, the solution should be treated gently to prevent breakage of the DNA strands.
Subsequent steps involve the separation of the DNA from other macromolecules in the lysate. Both phenol (that has been equilibrated with Tris buffer) and chloroform (with isoamyl alcohol as a de-foaming agent) are commonly used to dissociate protein from nucleic acids.
These reagents also remove lipids and some polysaccharides. Proteolytic enzymes such as pronase or proteinase K are often added to further remove protein. Proteinase K is a particularly useful enzyme; it is not denatured by SDS and in fact works more effectively in the presence of SDS.
The nucleic acids (including RNA) may then be precipitated in ice cold ethanol if the ionic strength of the solution is high. This is followed by RNase treatment to degrade the RNA. The solution may then be re-precipitated with ethanol.
In this precipitation, the ribonucleotides from RNase treatment will remain in solution leaving purified DNA in the pellet. The pellet can then be dissolved in an appropriate buffer.
Alcohol precipitations of DNA and RNA are widely used in molecular biology and are valuable because they allow the nucleic acids to be concentrated by removing them from solution as an insoluble pellet.
If concentrations of DNA are relatively high (> 1 μg/ml) DNA can be effectively precipitated in 10-15 minutes by shielding the negative charge with monovalent cations (0.3 M sodium or 2.5 M ammonium ions are commonly used) followed by the addition of 2 volumes of 95% ethanol.
A major consideration in any DNA isolation procedure is the inhibition or inactivation of DNases which can hydrolyze DNA. The buffer in which the cells are suspended should have a high pH (8.0 or greater) which is above the optimum of most DNases.
EDTA is also included in the re-suspension buffer to chelate divalent cations (such as Mg2+) which are required by DNases. The SDS also reduces DNase activity by denaturing these enzymes.
DNase activity is further controlled by keeping cells and reagents cold, using proteolytic enzymes such as pronase or proteinase K, and a heating step that will thermally denature DNase (but not hot enough to denature the DNA).
The procedure used here is useful for isolating DNA from a large variety of gram negative bacteria. It yields partially purified DNA of sufficient quality for most techniques, such as restriction digestion, ligation and cloning.
Further purification by additional solvent extraction may be required for experiments needing purer DNA (example – physical and chemical studies, such as melting curves).
1. Grow cells overnight in nutrient rich broth (LB broth).
2. Transfer 1.5 ml of culture to a micro-centrifuge tube and centrifuge at 10,000 rpm for 2 minutes. Collect the pellet and repeat with another 1.5 ml of culture containing cells. Drain the tubes on a paper towel briefly.
3. Re-suspend the pellet in 450 μl of TE buffer.
4. Add 45 μl of 10% SDS and 5 μl of 20 mg/ml proteinase K, mix and incubate for 1 hour at 37°C.
5. Add 500 μl phenol-chloroform and mix well by inverting the tube until the phases are completely mixed.
6. Centrifuge the mixture at 10,000 rpm in a microfuge for 2 minutes.
7. Transfer the upper aqueous phase to a new tube and re-extract by adding an equal volume (about 500 μl) of phenol-chloroform. Again centrifuge the mixture at 10,000 rpm in a microfuge for 5 minutes. Transfer the upper aqueous phase to a new tube.
8. Add 50 μl of sodium acetate and mix.
9. Add 300 μl of isopropanol and mix gently to precipitate the DNA.
10. Spool out the DNA with the help of an inoculation loop (or centrifuge at 10,000 rpm for 2-5 minutes).
11. Wash the DNA by dipping the end of the loop into 1 ml of 70% ethanol for 30 seconds and centrifuge briefly (or add 1 ml of 70% ethanol. Mix by inverting several times. Drain the tubes on a paper towel).
12. Re-suspend the DNA in 100-200 μl TE buffer (complete re-suspension may take several days).
13. Keep the DNA at 4°C for short-term and –20°C or –80°C for long-term storage.
Preparation of Solutions:
10% SDS (100 ml):
Dissolve 10 g SDS in 100 ml of distilled water. Heat gently to get SDS into solution.
Proteinase K (20 mg/ml):
Dissolve 20 mg proteinase K in 1 ml of distilled water.
Phenol-Chloroform (1:1):
Mix 50 ml of buffered phenol with 50 ml of chloroform.
3 M Sodium Acetate pH 5.2 (500 ml):
Dissolve 123 g in 450 ml of distilled water. Adjust the pH to 5.2 with glacial acetic acid. Make up to 500 ml.
3.
How to Isolate Yeast Genomic DNA?
The yeast, Saccharomyces cerevisiae, is a simple eukaryotic organism well suited as a tool for analysis of mammalian gene regulation. Development of yeast artificial chromosomes (YACs) greatly facilitated analysis of complex mammalian genetic loci by allowing cloning, maintenance, and manipulation of large stretches of exogenous DNA in yeast.
More recently, YACs have been used to generate transgenic mice. Introduction of mutations into YACs by homologous recombination coupled with the ability to produce transgenics provided a system to analyze the effect of the mutation in the context of the whole locus within an animal model.
1. Grow yeast in 100 ml culture medium for 8 hours.
2. Centrifuge the culture at 3000 rpm for 3 minutes.
3. Re-suspend the cells in 6 ml of distilled water. Aliquot into 1.5 ml microfuge tubes. Centrifuge at 3000 rpm for 3 minutes. Remove the supernatant.
4. Add 200 μl of isolation medium, 200 μl of phenol-chloroform and 0.3 g acid washed glass beads. Vortex for 2 minutes.
5. Add 200 μl TE pH 8.0 and centrifuge at 10,000 rpm for 5 minutes.
6. Preserve the supernatant and add 1 ml of 100% ethanol; mix by inversion. Centrifuge for 2 minutes at 10,000 rpm, remove the supernatant.
7. Reconstitute the pellet in 400 μl of TE pH 8.0 with 30 μg/ml RNase-A.
8. Incubate for 5 minutes at 37°C; add 40 μl 5M sodium acetate and 1ml ethanol and store at -20°C for a few hours.
9. Centrifuge for 10 minutes at 10,000 rpm and dissolve the DNA pellet in 25-50 μl of TE buffer.
Dissolve in 100 ml distilled water.
Add a pinch of streptomycin to the culture medium after sterilization.
Saboured dextrose broth or yeast peptone dextrose broth can also be used.
4. How to Isolate
Genomic DNA from Blood?
Blood contain a number of enzyme inhibitors that can interfere with isolation of DNA. Common anticoagulants such as heparin and EDTA can also interfere in DNA isolation. Erythrocytes (RBCs) from birds, fish and frogs contain nuclei and hence have genomic DNA, while those from mammals do not have nuclei.
Healthy mammalian blood contains approximately 1000 times more RBC- than WBC-containing nuclei—comprising leukocytes, lymphocytes, monocytes and granulocytes.
So it is better to remove all RBCs prior to DNA isolation to obtain higher yields. Selective lysis of erythrocytes using hypotonic solutions easily removes RBCs which are more susceptible to hypotonic buffers than WBCs. Alternatively Ficoll density-gradient centrifugation can be performed to recover mononuclear cells (lymphocytes and monocytes) and remove RBCs.
Cells without cell walls, like animal cells including blood cells, can be lysed efficiently by chemical or physical means. A common chemical method is to use a detergent to lyse the cells.
A detergent applied to cells will dissolve the fatty and proteinaceous cell membrane, so that the cells are lysed. A simple physical method for lysing cells is boiling. This also disrupts the cell membrane, lysing the cells.
The samples stored in deep freezers in EDTA vacutainer tubes are thawed, standard citrate buffer is added, mixed, and the tubes are centrifuged. The top portion of the supernatant is discarded and additional buffer is added, mixed, and the tube is again centrifuged. After the supernatant is discarded, the pellet is re-suspended in a solution of SDS detergent and proteinase K, and the mixture is incubated at 55°C for one hour.
The sample then is phenol extracted once with a phenol-chloroform-isoamyl alcohol solution, and after centrifugation the aqueous layer is removed to a fresh micro-centrifuge tube.
The DNA is ethanol precipitated, re-suspended in buffer, and then ethanol precipitated a second time. The pellet is dried, buffer is added and the DNA is re-suspended by incubation at 55°C overnight.
1. Add 0.8 ml of IX SSC buffer to 1 ml of sample, and mix.
2. Centrifuge for 1 minute at 12,000 rpm in a microfuge and discard 1 ml of supernatant.
3. Add 1 ml of IX SSC buffer again, vortex, and centrifuge at 12,000 rpm for 1 minute, and remove all of the supernatant.
4. Add 370 μl of 0.2 M sodium acetate to each pellet and mix by inverting the tubes briefly.
5. Add 25 μl of 10% SDS and 5 μl of proteinase K (20 mg/ml), mix by inverting the tubes briefly and incubate for 1 hour at 55°C.
6. Add 100 μl phenol-chloroform-isoamyl alcohol and mix for 30 seconds.
7. Centrifuge for 2 minutes at 12,000 rpm in a micro-centrifuge tube.
8. Collect the aqueous layer (top layer) carefully in a new micro-centrifuge tube, add 1 ml of cold 100% ethanol, mix, and incubate for 15 minutes at -20°C.
9. Centrifuge for 2 minutes at 12,000 rpm in a micro-centrifuge. Remove the supernatant and drain the tubes.
10. Add 180 μl 1X TE buffer, vortex, and incubate at 55°C for 10 minutes.
11. Add 20 μl 2 M sodium acetate and mix.
12. Add 500 μl of cold 100% ethanol, mix. Centrifuge for 1 minute at 12,000 rpm in a micro centrifuge.
13. Decant the supernatant and rinse the pellet with 1 ml of 70% ethanol. Centrifuge for 1 minute at 12,000 rpm in a micro-centrifuge.
14. Decant the supernatant, and dry the pellet in a Speed-Vac for 10 minutes, or until dry at room temperature.
15. Re-suspend the pellet by adding 200 μl of 1X TE buffer. Incubate overnight at 55°C, vortexing periodically to dissolve the genomic DNA.
16. Store the samples at -20°C.
Dissolve in 80 ml distilled water. Adjust the pH to 7.0 with concentrated HCl.
Make up the volume to 100 ml.
0.2 M Sodium Acetate (100 ml):
Dissolve 2.74 g sodium acetate in 80 ml distilled water.
Adjust the pH to 5.2 with glacial acetic acid.
Make up the volume to 100 ml
10 % SDS (50 ml):
Dissolve 5 g in 50 ml distilled water.
Proteinase K:
Dissolve 20 mg in 1 ml distilled water.
5. How to Isolate
DNA from Animal Cells?
To harvest, centrifuge cells from cell cultures, remove supernatant and then store the cells at -20°C or -80°C. These cells can be efficiently lysed using lysis buffer and proteinase K. Mechanical disruption using a homogenizer or mortar and pestle prior to lysis can reduce the lysis time.
1. Collect animal cells by centrifugation at 2000 rpm for 10 minutes at 4°C.
2. Re-suspend the cell pellet in cold cell lysis buffer (approximately 108 cells/ml).
3. Homogenize the cells in a glass homogenizer with a loose fitting pestle.
4. Centrifuge at 4000 rpm for 20 minutes at 4°C to pellet the nuclei.
5. Re-suspend the pellet in 8 ml of EDTA-NaCl, and add 0.8 ml of 10% or 1/10th the volume of cell lysis buffer from step 2.
6. Mix briefly by vortexing.
7. Add 50 μl of the proteinase K solution (10 mg/ml stock) and incubate at 37°C for 3-5 hours.
8. Add 0.5 ml of 5 M sodium acetate (or potassium acetate) and 8 ml of phenol-chloro- form-isoamyl alcohol. Mix gently by inverting the tube for 1 minute.
9. Centrifuge at 12,000 rpm for 10 minutes at 4°C.
10. Collect the upper aqueous phase.
11. Add an equal volume of chloroform-isoamyl alcohol. Mix gently by inverting the tube for 1 minute.
12. Centrifuge at 12,000 rpm for 10 minutes at 4°C. Collect the upper aqueous phase.
13. Add 2 volumes of 100% ethanol to precipitate the DNA.
14. Spool out the DNA with the inoculation loop. Dissolve in 1 ml of TE buffer.
15. Add 100 μl of 20X SSC and 10 μl of DNase-free RNase-A. Incubate for 1 hour at 37°C.
16. Extract the solution two times with chloroform-isoamyl alcohol.
17. Precipitate the DNA by adding 2 volumes of 100% ethanol.
18. Centrifuge at 5,000 rpm for 5 minutes at 4°C.
19. Wash the DNA pellet with 70% ethanol. Air dry briefly.
20. Dissolve the DNA in 200 μl of TE buffer.
Note – Do not over-dry the DNA pellet. It will be very difficult to dissolve the DNA in TE buffer.
6. How to Isolate
Genomic DNA from Eukaryotic Tissues?
Freshly collected tissues can be stored immediately at -20°C or-80°C, or in liquid nitrogen. Animal and human tissues can also be fixed in alcohol and formalin. However, long term storage of tissues in formalin will result in chemical modification of DNA.
These tissues can be effectively lysed using lysis buffer and proteinase K. Fresh and frozen tissues can be cut into pieces to aid lysis. Skeletal muscle, heart and skin tissues have an abundance of contractile proteins, connective tissue and collagen and care should be taken to ensure complete digestion with proteinase K. The method is also suitable for insect tissues, prawn and fish samples, and a wide variety of eukaryotic tissues.
1. Mince the tissue quickly and freeze in liquid nitrogen.
2. Immediately grind with pre-chilled mortar and pestle to a fine powder. Suspend in 1 ml digestion buffer per 100 mg of tissue.
3. Incubate samples with occasional shaking, in tightly capped microtubes, 30 to 60 minutes at 50°C.
4. Extract samples with an equal volume of phenol-chloroform-isoamyl alcohol.
5. Centrifuge for 10 minutes at 2000 rpm.
6. If phases do not resolve well, add another volume digestion buffer, omitting proteinase K, and repeat centrifugation. (If thick white material appears at interface, repeat organic extraction).
7. Transfer top aqueous layer to a new tube.
8. Add 0.5 volume of 7.5 M ammonium acetate and 2 volumes of 100% ethanol.
9. Centrifuge for 2 minutes at 2000 rpm (optional: to prevent shearing of high molecular weight DNA, remove organic solvents and salt by two dialyses against 100 volume TE buffer for > 24 hours; omit last step).
10. Wash with 70% ethanol, air dry, and re-suspend in TE buffer at 1mg/ml.
11. Remove residual RNA by adding 0.1% SDS and 1 μg/ml DNase-free RNase, incubating 1 hour at 37°C, and repeating ammonium acetate and ethanol precipitation of DNA and centrifugation.
7. How to Isolate
Plant DNA using CTAB Extraction Method?
Isolation of DNA from plant material presents special challenges and commonly used techniques often require adaptations before they can be used with plant samples. Several plant metabolites have chemical properties similar to those of nucleic acids, and are difficult to remove from DNA preparations.
Each plant is different and the cells of each kind of plant often contain special compounds, which must be extracted away from the DNA. Some plants, the cells of the tissue contain mucilage.
Mucilage is a very sticky, jelly-like substance whose function in the plant cells is not known for certain, but it is thought to act as antifreeze. Chemically, mucilage is a highly complex polysaccharide—very big, linear molecules, which have the ability to form large complexes with other large molecules such as DNA.
The mucilage is contained within the cytoplasm of the cell, while the DNA is contained within the nucleus (and within mitochondria and chloroplasts). To isolate DNA from the tissue of plants, the mucilage must be prevented from coming into contact with the DNA and complexing with it. In order to accomplish this, the DNA must remain within the nuclei while the mucilage is released from the cell and then poured off.
In this method the charged DNA molecules are initially dissolved in the polar salt-water CTAB solution. When this solution is mixed with chloroform, the non-polar compounds in the mixture dissolve into the chloroform layer, away from the aqueous DNA layer.
Subsequently, when the weakly polar isopropyl alcohol is added to the aqueous solution, the large negatively charged DNA molecules are no longer soluble in the solution, and they precipitate out as visible strands.
1. Chill a homogenizer with liquid nitrogen or dry ice.
2. Grind plant tissue to a fine powder using liquid nitrogen and transfer the frozen tissue to the tubes.
3. Add warm CTAB extraction buffer (maintained at 65°C) to the pulverized tissue and mix to wet thoroughly.
4. Incubate 10 to 60 minutes at 65°C with occasional mixing.
5. Extract the homogenate with an equal volume of chloroform-isoamyl alcohol. Mix well by inversion.
6. Centrifuge for 5 minutes at 8000 rpm at 4°C. Collect the upper aqueous phase.
7. Add 1/10 volume of CTAB-NaCl solution to the aqueous phase and mix well by inversion.
8. Extract with an equal volume of chloroform-isoamyl alcohol.
9. Mix, centrifuge and collect the upper phase.
10. Add 1 volume of CTAB precipitation solution.
11. Mix well by inversion. If precipitate is visible, proceed to next step. If not, incubate mixture for 30 minutes at 65°C.
12. Centrifuge for 5 minutes at 1000 rpm at 4°C.
13. Re-suspend the pellet in high-salt TE buffer. If the pellet is difficult to re-suspend, incubate 30 minutes at 65°C. Repeat until all or most of the pellet is dissolved (0.5 to 1 ml per gram of starting material).
14. Precipitate the nucleic acid by adding 0.6 volume isopropanol. Mix well.
15. Centrifuge for 15 minutes at 8000 rpm at 4°C.
16. Wash the pellet with 70% ethanol, dry, and re-suspend in a minimal volume of TE buffer.
17. Remove residual RNA by adding 0.1 % SDS and 1 μg/ml DNase-free RNase by incubating at 37°C for 1 hour.
18. Repeat CTAB-NaCl and ethanol precipitation of DNA.
Allow some time for the CTAB to dissolve and heat it to 65°C. The buffer is usually pH 8.0 without any adjustment. Do not autoclave CTAB buffer. Add 2-mercaptoethanol to the required amount of CTAB extraction buffer to give a final concentration of 2% (v/v).
8.
How to Isolate Chloroplast DNA?
In all terrestrial plants and algae, chloroplast DNA has been found to exist as single circular molecules ranging in molecular weight from 80 to 300 kb. The chloroplast genome is densely packed with genes and their functions are identified. They are involved in photosynthesis or occur as components of the chloroplast protein synthesizing system.
The physiological state of the starting leaf material is absolutely crucial to the success of a chloroplast DNA extraction. Wherever possible, only the freshest, youngest, and healthiest green leaves should be used.
It is far better to replant and wait for new growth than to extract from leaves that are beginning to yellow and senesce, or that have been stressed during growth, or that have wilted one or more times.
Some plants grown under high light intensities accumulate within their chloroplasts very high levels of starch that are very difficult to deplete, even by prolonged dark treatment.
This results in highly damaged chloroplasts and low DNA yields. In these cases the best solution is to grow the plants under moderate or low light intensities, example, under a greenhouse bench rather than on top of it.
If obtaining intact chloroplasts is difficult, incorporate 10%-25% (w/v) PEG 4000 in the homogenization buffer and all subsequent rinse buffers. Polyvinylpyrrolidone (PVP) at a concentration of 0.1% (w/v) in the homogenization buffer can also serve an effective adsorbent for tannins and other secondary plant compounds. Addition of 0.1% (w/v) antifoam to the homogenization buffer is useful with plant material that foams excessively during homogenization.
1. Keep the plants in the dark for 48 hours to reduce chloroplast starch levels.
2. Collect young and healthy leaves.
3. Wash leaves thoroughly in tap water and cool them to 0°C.
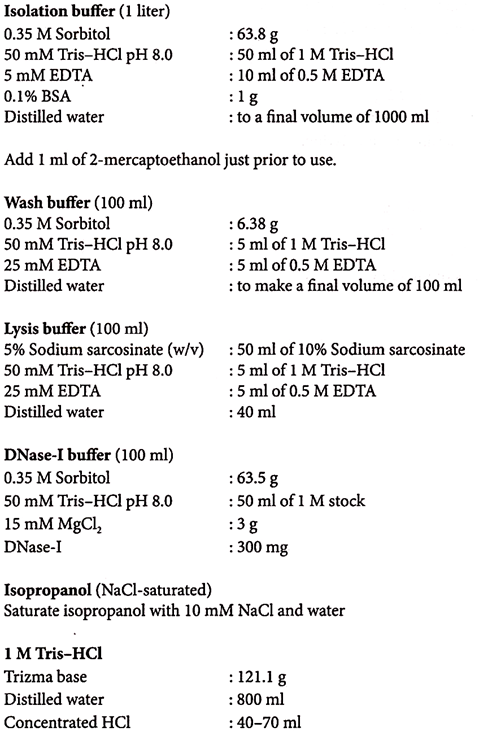
4. Place 1 g of leaves (cut into small pieces) in 4 ml of ice-cold isolation buffer.
5. Homogenize in a pre-chilled mortar and pestle for few minutes.
6. Filter the homogenate through dense nylon mesh (50 μM).
7. Centrifuge at 1000 rpm for 15 minutes at 4°C.
8. Collect the supernatant and re-centrifuge for 20 minutes at 4000 rpm.
9. Re-suspend the pellet in 400 μL wash buffer (for 1 g starting material) by vigorous swirling of the tubes.
10. Centrifuge at 4000 rpm for 20 minutes. Collect the chloroplast pellet.
Isolation of Chloroplast DNA by DNase-l Treatment Method:
1. Re-suspend the chloroplast pellet in 1 ml of DNase-I buffer (for 10 g starting material).
2. Incubate on ice for 1 hour.
3. Add three volumes of wash buffer and centrifuge at 2500 rpm for 15 minutes at 4°C.
4. Re-suspend pellet in wash buffer. Repeat washing and centrifuging for two times.
5. After final wash re-suspend pellet in 0.1 to 2.0 ml of wash buffer.
6. Add one-tenth volume of pronase (10 mg/ml self-digested for 2 hours at 37°C) and incubate for 2 minutes at room temperature.
7. Gently add one-fifth volume of lysis buffer and mix by slowly inverting the tube several times over a period of 10-15 minutes at room temperature.
8. Remove residual starch and cell wall debris from the chloroplast lysate by centrifuging at 10,000 rpm for 10 minutes at room temperature.
9. Collect the supernatant. Mix intensely for 5 minutes with an equal volume of buffer equilibrated phenol.
10. Centrifuge for 10 minutes at 12,000 rpm. Collect the upper aqueous phase.
11. Add equal volume of phenol-chloroform (1:1) and repeat step 10.
12. Collect the upper aqueous phase. Add equal volume of chloroform-isoamyl alcohol (24:1) and repeat the centrifugation step.
13. Precipitate the DNA with 1/10 volume of 5 M ammonium acetate and 1 volume of isopropanol at -20°C for 2-3 hours or overnight.
14. Centrifuge for 3-5 minutes at 12,000 rpm.
15. Wash the pellet with 70% ethanol, air dry and dissolve in 20 μl of TE buffer.
9. How to Isolate
Mitochondrial DNA?
Protocol:
Mitochondria Isolation:
1. Place 1 g of chilled leaves (cut into small pieces) or other tissues (animal origin) in 4 ml of ice-cold isolation buffer.
2. Homogenize in a pre-chilled mortar and pestle for few minutes.
3. Filter the homogenate through dense nylon mesh (50 μM).
4. Centrifuge at 1,000 rpm for 15 minutes at 4°C.
5. Collect the supernatant and re-centrifuge for 20 minutes at 4000 rpm.
6. Re-suspend the pellet in 400 μl rinse buffer (for 1 g starting material) by vigorous swirling of the tubes.
7. Centrifuge at 12,000 rpm for 20 minutes. Collect the mitochondria pellet.
Isolation of Mitochondrial DNA by DNase-l Treatment Method:
1. Re-suspend the pellet in 100 μl of DNase-I buffer (for 1 g starting material).
2. Incubate on ice for 1 hour.
3. Add three volumes of wash buffer and centrifuge at 2500 rpm for 15 minutes at 4°C.
4. Re-suspend pellet in wash buffer. Repeat washing and centrifuging twice.
5. After final wash re-suspend pellet in 0.1 to 2.0 ml of wash buffer.
6. Add one-tenth volume of pronase (10 mg/ml self-digested for 2 hours at 37°C) and incubate for 2 minutes at room temperature.
7. Gently add one-fifth volume of lysis buffer and mix by slowly inverting the tube several times over a period of 10-15 minutes at room temperature.
8. Remove residual starch and cell wall debris from the mitochondrial lysate by centrifuging at 10,000 rpm for 10 minutes at room temperature.
9. Collect the supernatant and mix intensely for 5 minutes with an equal volume of buffer equilibrated phenol.
10. Centrifuge for 10 minutes at 12,000 rpm. Collect the upper aqueous phase.
11. Add equal volume of phenol-chloroform (1:1) and repeat step 10.
12. Collect the upper aqueous phase. Add equal volume of chloroform-isoamyl alcohol (24:1) and repeat the centrifugation step.
13. Precipitate the DNA with 1/10 volume of 5 M ammonium acetate and 1 volume of isopropanol at -20°C for 2-3 hours or overnight.
14. Centrifuge for 3-5 minutes at 12,000 rpm.
15. Wash the pellet with 70% ethanol, air-dry and dissolve in 20 μI of TE buffer.
Preparation of Solutions: