The following are the common methods of DNA sequencing:- 1. Sequencing 2. Manual Sequencing 3. Automated Sequencing 4. Southern Blotting 5. Northern Blotting 6. Western Blotting.
Method # 1. Sequencing:
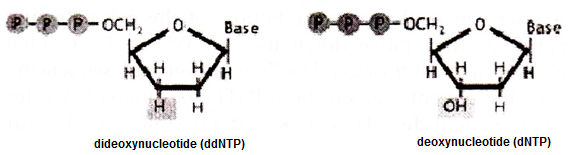
One of the major methods of DNA sequencing in known as chain termination sequencing, dideoxy sequencing, or Sanger sequencing after its inventor biochemist Frederick Sanger. The method is elegantly simple. While DNA chains are normally made up of deoxynucleotides (dNTPs), the Sanger method uses dideoxynucleotides.
Dideoxynucleotides (ddNTPs) are missing a hydroxy (OH) group at the 3′ position. This position is normally where one nucleotide attaches to another to form a chain. If there is no OH group in the 3′ position, the additional nucleotides cannot be added to the chain, thus interrupting chain elongation.
Method # 2. Manual Sequencing:
1. Begin the process by synthesizing a chain that is complimentary to the template you want to analyse.
2. Add a specific primer that you know will anneal.
3. Divide your sample among four test tubes. One test tube will be used for each specific nucleotide (dGTP, dATP, dCTP, and dTTP).
4. Add DNA polymerase to each tube and one specific nucleotide per tube.
5. Add ddNTPs to all four tubes. Once you add the ddNTPs, there is no way for the chain to keep elongating; hence you have dd chain termination.
6. Run this on a polyacrylamide gel using one lane per reaction tube (dGTP, dATP, dCTP, and dTTP).
7. To sequence, read the order of bases from the smallest to the largest.
Method # 3. Automated Sequencing:
An automated sequencer runs on the same principle as the Sanger method (dideoxynucleotide chain termination). A laser constantly scans the bottom of the gel, detecting bands as they move down the gel.
Where the manual method uses radioactive labelling, automated sequencing uses fluorescent tags on the ddNTPs (a different dye for each nucleotide). This makes it possible for all four reactions (dGTP, dATP, dCTP, and dTTP) to be run in one lane, so you can have huge numbers of reactions on one gel. This is a very efficient method.
It is important to remember, however, that a computer can make mistakes. Don’t trust the computer. Always check your printout for accuracy. You are looking for a good signal, at least in the 100s, and proper spacing, ideally about 12. Look also for big gaps between the bases since the computer can miss bases. It may often miss a G after an A, especially after an AA.
Method # 4. Southern Blotting:
Southern blotting was named after Edward M. Southern who developed this procedure at Edinburgh University in the 1970s. To oversimplify, DNA molecules are transferred from an agarose gel onto a membrane. Southern blotting is designed to locate a particular sequence of DNA within a complex mixture.
For example, Southern Blotting could be used to locate a particular gene within an entire genome. The amount of DNA needed for this technique is dependent on the size and specific activity of the probe. Short probes tend to be more specific. Under optimal conditions, you can expect to detect 0.1 pg of the DNA for which you are probing.
Method # 5. Northern Blotting:
Sometimes it’s a bit hard to understand, but there is humour in science. In the 1970s, E.M. Southern developed a method for locating a particular sequence of DNA within a complex mixture. This technique came to be known as Southern blotting.
In a tongue-in-cheek fashion, those who used a similar method for locating a sequence of RNA named it Northern blotting. It is also known as Northern hybridisation or RNA hybridisation. The procedure for and theory behind Northern blotting is almost identical to that of Southern blotting, except you are working with RNA instead of DNA.
Method # 6. Western Blotting:
Western blot analysis can detect one protein in a mixture of any number of proteins while giving you information about the size of the protein. It does not matter whether the protein has been synthesised in vivo or in vitro. This method is, however, dependent on the use of a high- quality antibody directed against a desired protein. So you must be able to produce at least a small portion of the protein from a cloned DNA fragment.
You will use this antibody as a probe to detect the protein of interest. Western blotting tells you how much protein has accumulated in cells. If you are interested in the rate of synthesis of a protein, Radio-Immune Precipitation (RIP) may be the best assay for you. Also, if a protein is degraded quickly, Western blotting won’t detect it well; you’ll need to use (RIP).